Programming with R: Guide
“Introduction to R and RStudio” - 45 minutes
Before Starting The Workshop
Please ensure you have the latest version of R and RStudio installed on your machine.
Introduction to RStudio
Why R?
- free
- open source
Why RStudio?
- provides a fully integrated environment
- built in version control and project management
- works on all platforms
Basic layout
three panels:
- The interactive R console (entire left)
- Environment/History (tabbed in upper right)
- Files/Plots/Packages/Help/Viewer (tabbed in lower right)
Editor panel when you open scripts
Work flow within RStudio
There are two main ways one can work within RStudio.
- Test and play within the interactive R console then copy code into a .R file to run later.
- Start writing in an .R file and use RStudio’s command / short cut to push current line, selected lines or modified lines to the interactive R console.
Run button and key shortcuts
- ctrl-enter windows/linux
- command-enter mac
Project managment
Mostly use the console
- run code and test commands
”>” cursor - similar to the shell
- type commands
- R executes
- returns result
The simplest thing you could do with R is do arithmetic:
1 + 100
Ignore the [1] for now.
Incomplete commands:
> 1 +
Can cancel with the “Esc” key, or “Ctrl+C” in command line - can use for running code too
order of operations
3 + 5 * 2
Use parentheses to force order:
(3 + 5) * 2
Use these to make code easier to read
(3 + (5 * (2 ^ 2))) ## hard to read
3 + 5 * 2 ^ 2 ## clear, if you remember the rules
3 + 5 * (2 ^ 2) ## if you forget some rules, this might help
”##” indicate comments
Scientific notation
2/10000
5e3 ## Note the lack of minus here
Mathematical functions
functions use name then parenthesis
sin(1) ## trigonometry functions
log(1) ## natural logarithm
log10(10) ## base-10 logarithm
exp(0.5) ## e^(1/2)
Can look up functions in google or use Autocomplete (tab)
Comparing things
1 == 1 ## equality (note two equals signs, read as "is equal to")
1 != 2 ## inequality (read as "is not equal to")
1 < 2 ## less than
1 <= 1 ## less than or equal to
1 > 0 ## greater than
1 >= -9 ## greater than or equal to
float point error, use all.equal instead of ‘==’ for non-integers
Variables and assignment
use assignment arrow to save values to variables
x <- 1/40
No output
x
stored as a decimal approximation called floating point number.
point out x in environment variables
log(x)
Can reassign values
x <- 100
Can reference the variable in the assignment
x <- x + 1 ##notice how RStudio updates its description of x on the top right tab
The right hand side of the assignment can be any valid R expression. The right hand side is fully evaluated before the assignment occurs.
character values:
y <- "green"
Variable names rules: can contain letters, numbers, underscores and periods cannot start with a number nor contain spaces at all.
Naming conventions:
- periods.between.words
- underscores_between_words
- camelCaseToSeparateWords
consistency is important
can use = operator for assignment:
z = 1/40
Less common, and sometimes it is confusing to use ‘=’ instead of ‘<-‘ remember consistency!
Functions
built in functions coding your own supplemental lesson
R Packages
- install packages:
install.packages("packagename") - make a package available for use:
library(packagename)
install.packages("gapminder")
text scrolling for install
library(gapminder)
Point out how to install via the Packages tab.
Other useful commands:
- You can see what packages are installed by typing
installed.packages() - You can update installed packages by typing
update.packages() - You can remove a package with
remove.packages("packagename")
CHALLENGES (allot 15 min)
“Seeking Help” - 20 minutes
Reading Help files
help(function_name)
?function_name
Each help page is broken down into sections:
- Description: An extended description of what the function does.
- Usage: The arguments of the function and their default values.
- Arguments: An explanation of the data each argument is expecting.
- Details: Any important details to be aware of.
- Value: The data the function returns.
- See Also: Any related functions you might find useful.
- Examples: Some examples for how to use the function.
Some may have different sections, but these are the main ones.
Help files make it easier to use R because you don;’t have to remember the usage of every function.
Special Operators
To seek help on special operators, use quotes:
?"+"
Getting help on packages
Many packages come with “vignettes”: tutorials and extended example documentation. Without any arguments
vignette() will list all vignettes for all installed packages;
vignette(package="package-name") will list all available vignettes for package-name
and vignette("vignette-name") will open the specified vignette.
If a package doesn’t have any vignettes, you can usually find help by typing
help("package-name").
When you kind of remember the function
fuzzy search:
??function_name
When you have no idea where to begin
CRAN Task View - show website: http://cran.at.r-project.org/web/views
When your code doesn’t work: seeking help from your peers
Stack Overflow - search using the [r] tag.
CHALLENGES - allot 10 min
“Data Structures” - finish at 10 am
Data Types in R
Review operators:
x + z
Try adding two different types of data:
x + y
Gives error because 101 + “green” is nonsense.
5 main data types: double, integer, complex, logical, and character.
typeof(3.14)
A double, also referred to as a floating point number, is how R stores numeric values
by default.
typeof(1L)
To use an integer value in R, we use the L to tell R that this value is an integer value.
Without the L, R would store this value as a double.
typeof(1+1i)
R can also support complex values as well. Unless you are doing mathematical analyses or
complicated transformations, chances are you will not encounter this data type very often.
typeof(TRUE)
[1] "logical"
Logical data types are particularly helpful in subsetting data frames and other types of data manipulation. We will explore this concept more later.
typeof('banana')
[1] "character"
Lastly, R stores strings as the character type.
No matter how complicated our analyses become, all data in R is interpreted as one of these basic data types.
Vectors
Remember the [1]? R never uses just a single value, but instead uses vectors. Output before was a vector of length 1.
As seen in the challenges, we can build vectors using the c function.
x <- c(2, 4, 6, 8, 10, 12, 14, 16)
x
colon operator quickly creates sequential vectors:
y <- 1:8
y
can specify whatever start and stop point we want
-4:7
[1] -4 -3 -2 -1 0 1 2 3 4 5 6 7
R is vectorized operations on the entire vector - returns a vector
y + 10
[1] 11 12 13 14 15 16 17 18
x * 2
[1] 4 8 12 16 20 24 28 32
x + y
[1] 3 6 9 12 15 18 21 24
when operating on two or more vectors, R performs the operation element by element:
x: 2 4 6 8 10 12 14 16
+ + + + + + + +
y: 1 2 3 4 5 6 7 8
--------------------------
3 6 9 12 15 18 21 24
Vectors can be made up of any of the basic data types.
Character Vectors:
a <- c("one", "two", "three", "four")
a
similar to typeof, str will tell us the data type.
also will give us a compact view of some basic information about our object.
str(a)
chr that this is a character vector numbers in the brackets indicates the dimensions of our vector. then a list the first few elements
Logical Vectors:
b <- c(TRUE, TRUE, FALSE, TRUE)
b
we can use c to add elements to an existing vector
c(x, 20, 25)
Changes won’t be saved until we use the assignment arrow
modify y to contain all numbers to 20 by adding on to the existing vector
y <- c(y, 9:20)
y
This is a nested operation, from Order of Opers. earlier the sequence inside the parenthesis is created first, then added to y
length can quickly return the length of a vector.
length(y)
Other data structures, lists and matrices
use ?list(), ?matrix() or supplemental lesson to learn about them.
primarily data frames today, continued after break:
CHALLENGES - allot 10 minutes
COFFEE BREAK - 15 min
Subsetting Data - 45 minutes
** HERE **
R’s power comes from it’s vectorization. many powerful subset operators that will allow you to easily perform complex operations on any kind of dataset without the resource depletion of loops.
Let’s start with a data structure we’ve seen before, the workhorse of R: atomic vectors.
x <- c(5.4, 6.2, 7.1, 4.8, 7.5)
can name elements within our vectors using the names function.
names(x) <- c('a', 'b', 'c', 'd', 'e')
x
how do we get to individual contents?
Accessing elements using their indices
we can give their corresponding index, starting from one:
x[1]
x[4]
the square brackets operator is a function For atomic vectors (and matrices), it means “get me the nth element”.
We can ask for multiple elements at once:
x[c(1, 3)]
Or slices of the vector:
x[1:4]
the : operator lets us select a range of elements
We can ask for the same element multiple times:
x[c(1,1,3)]
a number outside of the vector, R will return missing values
x[6]
This is a vector of length one containing an NA, whose name is also NA.
If we ask for the 0th element, we get an empty vector:
x[0]
named numeric(0)
R starts indices with 1 instead of 0 like other programming languages such as C and python
Skipping and removing elements
use a negative number to return every element except for the one specified:
x[-2]
We can skip multiple elements:
x[c(-1, -5)] ## or x[-c(1,5)]
Combining positive and negative indices will return an error.
To make the subset permanent we need to assign the value.
x <- x[-4]
x
CHALLENGE - allot 5 minutes
Subsetting by name
can extract elements by using their name
x[c("a", "c")]
more reliable since positions can change. But we cannot as easily skip or remove by name.
To skip (or remove) a single named element:
x[-which(names(x) == "a")]
The which function returns the indices of all TRUE elements of its argument.
Step by step analysis of command:
names(x) == "a"
The condition operator is applied to every name of the vector x. Only the
first name is “a” so that element is TRUE.
which then converts this to an index:
which(names(x) == "a")
Only the first element is TRUE, so which returns 1.
the ‘-‘ makes this index negative and removes the element
Skipping multiple named indices is similar, but uses a different comparison operator:
x[-which(names(x) %in% c("a", "c"))]
%in% goes through each element of its left argument, in this case the
names of x, and asks, “Does this element occur in the second argument?”.
why can’t we use == like before? Good question.
names(x) == c('a', 'c')
gives a warning
== works by comparing each element of its left argument
to the corresponding element of its right argument.
Here’s a mock illustration:
c("a", "b", "c", "d", "e") ## names of x
| | | | | ## The elements == is comparing
c("a", "c")
when one vector is shorter than the other, it gets recycled:
c("a", "b", "c", "d", "e") ## names of x
| | | | | ## The elements == is comparing
c("a", "c", "a", "c", "a")
R repeats c("a", "c") two and a half times. If the longer
vector length isn’t a multiple of the shorter vector length, then
R will also print out a warning message.
This difference between == and %in% is important to remember,
because it can introduce hard to find and subtle bugs!
CHALLENGES - allot 10 min
Using Logical Operations to Subset Data
We can subset data by using boolean vectors:
x[c(TRUE, TRUE, FALSE, FALSE, FALSE)]
R will return any values that are indicated by TRUE in your vector, and filter out any that
are FALSE.
x[c(TRUE, FALSE)]
R also recycled our logical vector
comparison operators evaluate to logical vectors we can use them to succinctly subset vectors
x > 7
nest our comparison inside of our subsetting operators to tell R to return a subset
x[x > 7]
Combining logical conditions
&, the “logical AND” operator: returnsTRUEif both the left and right areTRUE.|, the “logical OR” operator: returnsTRUE, if either the left or right (or both) areTRUE.
The recycling rule applies with both of these
&& and || do not use the recycling rule: they only look at the first element of each
vector and ignore the remaining elements. Usually used in programming not data analysis
!, the “logical NOT” operator: convertsTRUEtoFALSEandFALSEtoTRUE. can negate a single logical condition, or a whole vector of conditions
the all function returns TRUE if every element of the vector is TRUE
the any function returns TRUE if one or more elements of the vector are TRUE.
CHALLENGES allot 5 min
“Exploring Data Frames” - done at noon
Data Frames
so far data structures contained all of the same data type one of R’s most powerful features is its ability to deal with tabular data (like spreadsheet or CSV) Data Frames are built from vectors but can contain vectors of different data types.
build a data frame from existing vectors - data.frame() command
coat <- c("calico", "black", "tabby")
weight <- c(2.1, 5.0, 3.2)
likes_string <- c(1,0,1)
cats <- data.frame(coat, weight, likes_string)
cats
can pull out columns by specifying them using the $ operator
cats$weight
cats$coat
can perform operations on columns within our data frame, just like with vectors
#### Say we discovered that the scale weighs two Kg light:
cats$weight + 2
paste("My cat is", cats$coat)
CHALLENGES allot 5 min
add an additional column for age using c
age <- c(2,3,5,12)
cats
We can then add this as a column in our data frame by using the cbind() function:
cats <- cbind(cats, age)
Error - there are more elements in age but only 3 rows in cats.
age <- c(4,5,8)
cats <- cbind(cats, age)
cats
add a row, rows are lists since they contain different types of elements:
newRow <- list("tortoiseshell", 3.3, TRUE, 9)
cats <- rbind(cats, newRow)
Our list had the correct number of elements, so why did R give us a warning?
class(cats$coat)
Factors are data classes that R uses to handle categorical data. categories are called levels
Anything new that doesn’t fit into one of its categories is rejected as nonsense and
is replaced by an NA until we explicitly add that as a level in the factor:
levels(cats$coat)
levels(cats$coat) <- c(levels(cats$coat), 'tortoiseshell')
cats <- rbind(cats, list("tortoiseshell", 3.3, TRUE, 9))
can also change the column to character to prevent this
str(cats)
cats$coat <- as.character(cats$coat)
str(cats)
We can now add rows and columns, but we’ve accidentally added a garbage row:
cats
coat weight likes_string age
1 calico 2.1 1 4
2 black 5.0 0 5
3 tabby 3.2 1 8
4 <NA> 3.3 1 9
5 tortoiseshell 3.3 1 9
We can ask for a data.frame minus this offending row:
cats[-4,]
Notice the comma with nothing after it to indicate we want to drop the entire fourth row.
using na.omit allows us to drop all rows with NA values:
na.omit(cats)
Let’s reassign the output to cats, so that our changes will be permanent:
cats <- na.omit(cats)
remember that columns are vectors or factors, and rows are lists.
We can also glue two dataframes together with rbind:
cats <- rbind(cats, cats)
cats
But now the row names are unnecessarily complicated. We can remove the rownames, and R will automatically re-name them sequentially:
rownames(cats) <- NULL
cats
CHALLENGES - allot 5 min
let’s use a more realistic dataset
the gapminder data set built into the gapminder package.
install the gapminder package if you havent already
install.packages("gapminder")
load it now using the library command (explain how to use the packages tab
library('gapminder')
to make our analysis reproducible, we should put the code into a script file
mention loading libraries in script files
investigate the data - check
out what the data looks like with str:
str(gapminder)
We can also examine individual columns of the data.frame with our typeof function:
typeof(gapminder$year)
typeof(gapminder$lifeExp)
typeof(gapminder$country)
str(gapminder$country)
information about its dimensions;
remembering that str(gapminder) said there were 1704 observations of 6
variables in gapminder
what do you think the following will produce, and why?
length(gapminder)
try it, point out that data frames are lists of vectors - each column is a vector/factor and each row is a list (hence the different data types)
typeof(gapminder)
length gave us 6 because gapminder is built out of a list of 6
columns
To get the number of rows and columns in our dataset, try:
nrow(gapminder)
ncol(gapminder)
Or, both at once:
dim(gapminder)
titles of all the columns:
colnames(gapminder)
ask ourselves if the structure R is reporting matches our intuition or expectations do the basic data types reported for each column make sense? No? We need to fix it now
Once we’re happy that the data types and structures seem reasonable start digging into our data proper
head(gapminder)
CHALLENGES - 5 min
Subsetting Data Frames
data frames are lists of vectors, so selecting a single element returns a single vector, or column of the data frame.
head(gapminder[5])
the c command to returns multiple columns:
head(gapminder[c(1,5)])
$ provides a convenient shorthand to extract columns by name:
head(gapminder$year)
With two arguments, [ subsets on typical matrix format
if one of the arguments is blank, R will
default to include all of the rows or columns:
gapminder[1:3,]
If we subset a single row, the result will be a data frame
gapminder[3,]
But for a single column the result will be a vector.
CHALLENGES - 15 min
“Lunch” - 1 hour
Control Flow - 35 minutes
Often when we’re coding we want to control the flow of our actions. This can be done by setting actions to occur only if a condition or a set of conditions are met. Alternatively, we can also set an action to occur a particular number of times.
There are several ways you can control flow in R. For conditional statements, the most commonly used approaches:
## if
if (condition is true) {
perform action
}
## if ... else
if (condition is true) {
perform action
} else { ## that is, if the condition is false,
perform alternative action
}
if we want R to print a message if a variable x has a particular value:
## sample a random number from a Poisson distribution
## with a mean (lambda) of 8
x <- rpois(1, lambda=8)
if (x >= 10) {
print("x is greater than or equal to 10")
}
x
you may not get the same output as your neighbour
Let’s set a seed so that we all generate the same ‘pseudo-random’ number
set.seed(10)
x <- rpois(1, lambda=8)
if (x >= 10) {
print("x is greater than or equal to 10")
} else if (x > 5) {
print("x is greater than 5")
} else {
print("x is less than 5")
}
In the above case, the function rpois() generates a random number following a
Poisson distribution with a mean (i.e. lambda) of 8. The function set.seed()
guarantees that all machines will generate the exact same ‘pseudo-random’
number
when R evaluates the condition inside if() statements, it is
looking for a logical element
This can cause some headaches for beginners.
x <- 4 == 3
if (x) {
"4 equals 3"
}
As we can see, the message was not printed because the vector x is FALSE
x <- 4 == 3
x
CHALLENGES - 5 min
Did anyone get a warning message like this?
Warning in if (gapminder$year == 2012) {: the condition has length > 1 and
only the first element will be used
If your condition evaluates to a vector with more than one logical element,
the function if() will still run, but will only evaluate the condition in the first
element. Here you need to make sure your condition is of length 1.
The any() function will return TRUE if at least one
TRUE value is found within a vector, otherwise it will return FALSE.
This can be used in a similar way to the %in% operator.
The function all(), as the name suggests, will only return TRUE if all values in
the vector are TRUE.
While loops
to repeat an operation until a certain condition is met use a while() loop
while(this condition is true){
do a thing
}
here’s a while loop
that generates random numbers from a uniform distribution (the runif function)
between 0 and 1 until it gets one that’s less than 0.1.
z <- 1
while(z > 0.1){
z <- runif(1)
print(z)
}
You have to be careful that you don’t end up in an infinite loop because your condition is never met.
Repeating operations
If you want to iterate over
a set of values a for() loop will do the job.
This is the most
flexible of looping operations, but therefore also the hardest to use
correctly.
Avoid using for() loops unless the the calculation at each iteration depends on the results of previous iterations.
The basic structure of a for() loop is:
for(iterator in set of values){
do a thing
}
For example:
for(i in 1:10){
print(i)
}
The 1:10 bit creates a vector on the fly; you can iterate
over any other vector as well.
We can use a for() loop nested within another for() loop to iterate over two things at
once.
for(i in 1:5){
for(j in c('a', 'b', 'c', 'd', 'e')){
print(paste(i,j))
}
}
Rather than printing the results, we could write the loop output to a new object.
output_vector <- c()
for(i in 1:5){
for(j in c('a', 'b', 'c', 'd', 'e')){
temp_output <- paste(i, j)
output_vector <- c(output_vector, temp_output)
}
}
output_vector
This approach can be useful, but ‘growing your results’ is computationally inefficient, so avoid it when you are iterating through a lot of values.
A better way is to define your (empty) output object before filling in the values.
output_matrix <- matrix(nrow=5, ncol=5)
j_vector <- c('a', 'b', 'c', 'd', 'e')
for(i in 1:5){
for(j in 1:5){
temp_j_value <- j_vector[j]
temp_output <- paste(i, temp_j_value)
output_matrix[i, j] <- temp_output
}
}
output_vector2 <- as.vector(output_matrix)
output_vector2
CHALLENGES - 15 min
Dataframe Manipulation with dplyr - finish by 2:30
Manipulation of dataframes means many things to many researchers, we often select certain observations (rows) or variables (columns), we often group the data by a certain variable(s), or we even calculate summary statistics. We can do these operations using the normal base R operations:
mean(gapminder[gapminder$continent == "Africa", "gdpPercap"])
mean(gapminder[gapminder$continent == "Americas", "gdpPercap"])
mean(gapminder[gapminder$continent == "Asia", "gdpPercap"])
This can be repetitive, and repetition will cost you time, make your code bulky and hard to read and potentially introduce some nasty bugs
The dplyr package
the dplyr package provides a number of useful functions for manipulating dataframes
in a way that will reduce the above repetition, reduce the probability of making
errors, and probably even save you some typing. the dplyr grammar can also make your code easier to read.
we’re going to cover 5 of the most commonly used functions as well as using
pipes (%>%) to combine them
select()filter()group_by()summarize()mutate()
If you have have not installed this package earlier, please do so:
install.packages('dplyr')
Now let’s load the package:
library(dplyr)
Using select()
If we wanted to use only a few of the variables in
our dataframe we could use the select() function.
year_country_gdp <- select(gapminder,year,country,gdpPercap)
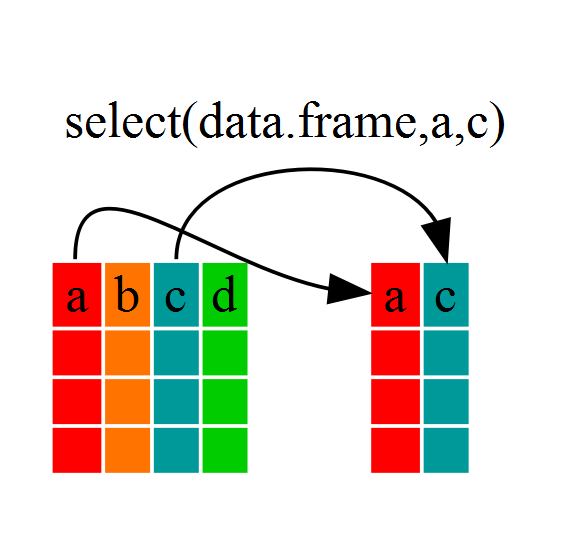
look at year_country_gdp
see that it only contains the year, country and gdpPercap.
we used ‘normal’ grammar
but the strengths of dplyr lie in combining several functions using pipes. Since the pipes grammar
is unlike anything we’ve seen in R before, let’s repeat what we’ve done above
using pipes.
year_country_gdp <- gapminder %>% select(year,country,gdpPercap)
To help you understand why we wrote that in that way, let’s walk through it step
by step
First we summon the gapminder dataframe and pass it on, using the pipe
symbol %>%, to the next step, which is the select() function.
In R, a pipe symbol is %>% while in the
shell it is |
Using filter()
using the above but only with European
countries, we can combine select and filter
year_country_gdp_euro <- gapminder %>%
filter(continent=="Europe") %>%
select(year,country,gdpPercap)
line breaks between specific portions of our command make the code easier to read
first we pass the data frame to filter then pass the filtered dataframe to select.
If we reversed this, it would not work since we removed the continent data with select
CHALLENGES - 5 min
Using group_by() and summarize()
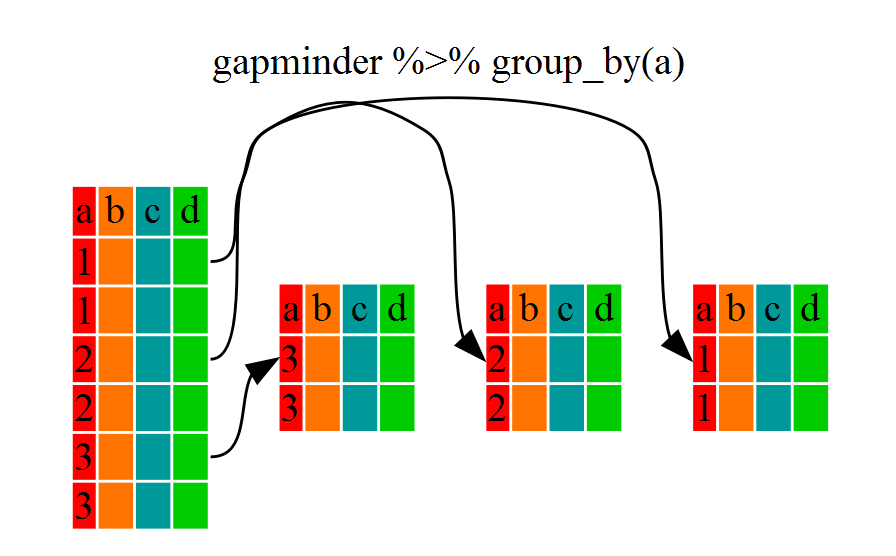
If we want to do the same above for each country, we can reduce repetitiveness with group_by
group_by will essentially use every unique criteria that you
could have used in filter.
str(gapminder)
str(gapminder %>% group_by(continent))
notice that the structure of the dataframe where we used group_by()
(grouped_df) is not the same as the original gapminder (data.frame).
A
grouped_df can be thought of as a list where each item in the listis a
data.frame which contains only the rows that correspond to the a particular
value.
Using summarize()
group_by() is much more
exciting in conjunction with summarize()
summarize allows us to create new
variable(s) by using functions that repeat for each of the continent-specific
data frames
gdp_bycontinents <- gapminder %>%
group_by(continent) %>%
summarize(mean_gdpPercap=mean(gdpPercap))
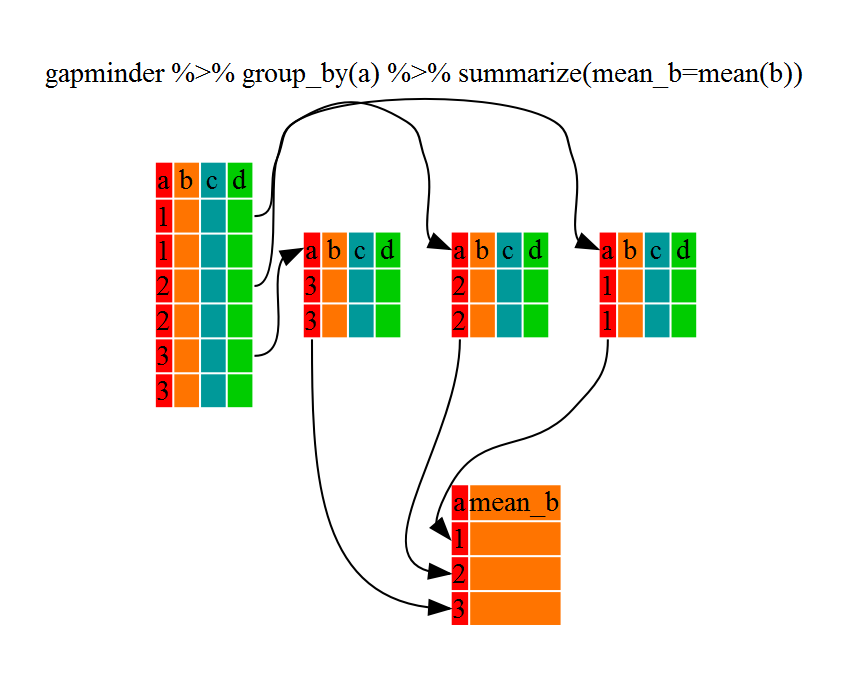
That allowed us to calculate the mean gdpPercap for each continent, but it gets even better.
CHALLENGES - 5 min
group_by() allows us to group by multiple variables. Let’s group by year and continent.
gdp_bycontinents_byyear <- gapminder %>%
group_by(continent,year) %>%
summarize(mean_gdpPercap=mean(gdpPercap))
we can define more than 1 variable with summarize
gdp_pop_bycontinents_byyear <- gapminder %>%
group_by(continent,year) %>%
summarize(mean_gdpPercap=mean(gdpPercap),
sd_gdpPercap=sd(gdpPercap),
mean_pop=mean(pop),
sd_pop=sd(pop))
Using mutate()
We can create new variables prior to (or even after) summarizing information using mutate().
gdp_pop_bycontinents_byyear <- gapminder %>%
mutate(gdp_billion=gdpPercap*pop/10^9) %>%
group_by(continent,year) %>%
summarize(mean_gdpPercap=mean(gdpPercap),
sd_gdpPercap=sd(gdpPercap),
mean_pop=mean(pop),
sd_pop=sd(pop),
mean_gdp_billion=mean(gdp_billion),
sd_gdp_billion=sd(gdp_billion))
** CHALLENGES ** - allot 10 min
“Coffee Break” - 15 min
Creating Publication-Quality Graphics - 1 hour and 15 min
Plotting our data is one of the best ways to quickly explore it and the various relationships between variables.
Today we’ll be learning about the ggplot2 package, because it is a bit harder to learn but produces better looking plots than the base plotting ability.
ggplot2 is built on the grammar of graphics (where the gg comes from) the idea that any plot can be expressed from the same set of components:
- a data set
- a coordinate system
- a set of geoms–the visual representation of data points.
The key to understanding ggplot2 is thinking about a figure in layers.
Let’s start off with an example. The first thing we need to do is load the ggplot2 package
library("ggplot2")
If you haven’t previously installed the package, install it now using the command install.packages("ggplot2"). Then load it using the command above.
to begin graphing, we use the ggplot function
this lets R
know that we’re creating a new plot, and any of the arguments we give the
ggplot function are the global options for the plot: they apply to all
layers on the plot.
library("ggplot2")
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point()
mention that line breaks can be anywhere, but you need to have the + at the end of the line.
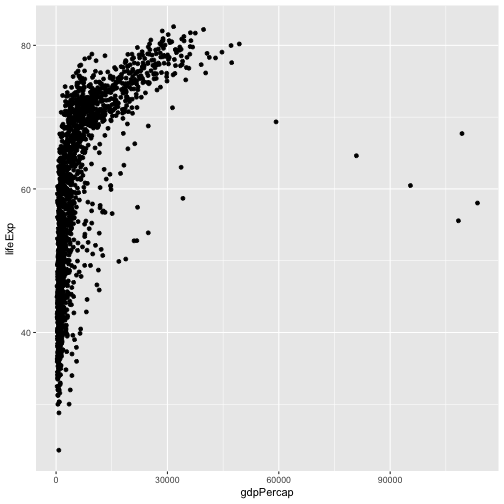
We’ve passed in two arguments to ggplot.
First, we tell ggplot what data we
want to show on our figure
For the second argument we passed in the aes function, which
tells ggplot how variables in the data map to aesthetic properties of
the figure, in this case the x and y locations.
Here we told ggplot we
want to plot the “gdpPercap” column of the gapminder data frame on the x-axis, and
the “lifeExp” column on the y-axis.
we didn’t need to explicitly
pass aes these columns (e.g. x = gapminder[, "gdpPercap"])
Other options that can be set with the aes function include color, size, transparency and shape. We will talk more about that later.
By itself, the call to ggplot isn’t enough to draw a figure:
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp))
We need to tell ggplot how we want to visually represent the data, which we
do by adding a new geom layer. In our example, we used geom_point, which
tells ggplot we want to visually represent the relationship between x and
y as a scatterplot of points:
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point()
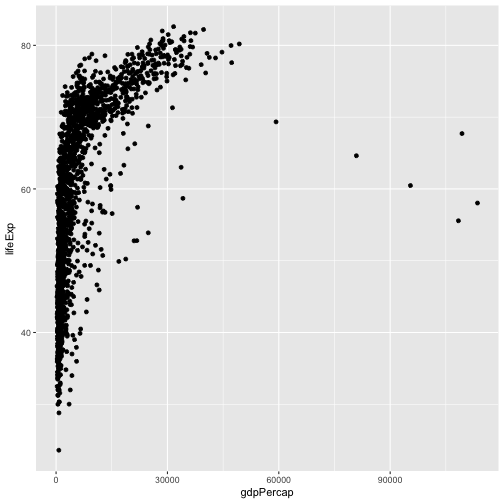
CHALLENGES - 10 min
Layers
Using a scatterplot probably isn’t the best for visualizing change over time.
Instead, let’s tell ggplot to visualize the data as a line plot:
ggplot(data = gapminder, aes(x=year, y=lifeExp, by=country, color=continent)) +
geom_line()
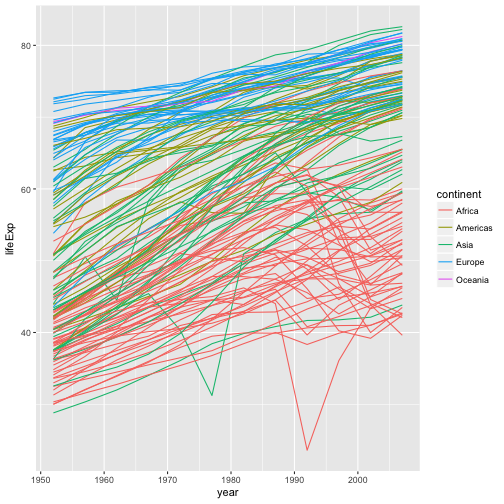
Instead of adding a geom_point layer, we’ve added a geom_line layer. We’ve
added the by aesthetic, which tells ggplot to draw a line for each
country.
if we want to visualize both lines and points on the plot? We can simply add another layer to the plot:
ggplot(data = gapminder, aes(x=year, y=lifeExp, by=country, color=continent)) +
geom_line() + geom_point()
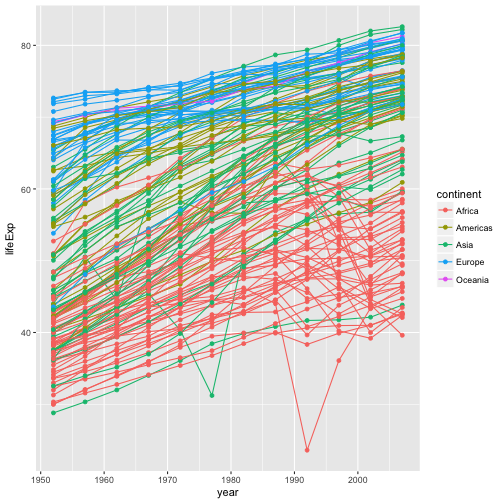
note that each layer is drawn on top of the previous layer.
ggplot(data = gapminder, aes(x=year, y=lifeExp, by=country)) +
geom_line(aes(color=continent)) + geom_point()
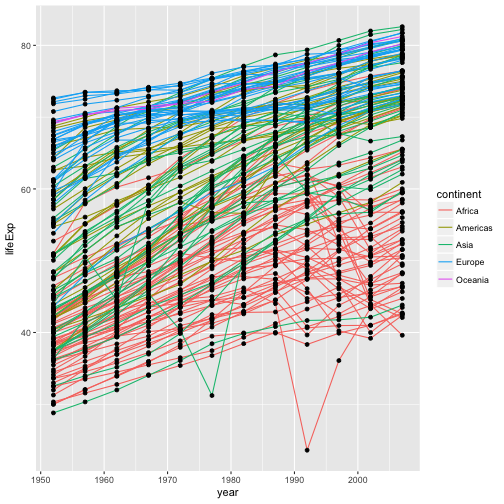
the aesthetic mapping of color has been moved from the
global plot options in ggplot to the geom_line layer so it no longer applies
to the points.
Now we can clearly see that the points are drawn on top of the lines.
Demonstrate changing an aesthetic to a solid color:
ggplot(data = gapminder, aes(x=year, y=lifeExp, by=country)) +
geom_line(aes(color="red")) + geom_point()
Transformations and statistics
ggplot also makes it easy to overlay statistical models over the data. To
demonstrate we’ll go back to our first example:
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point()
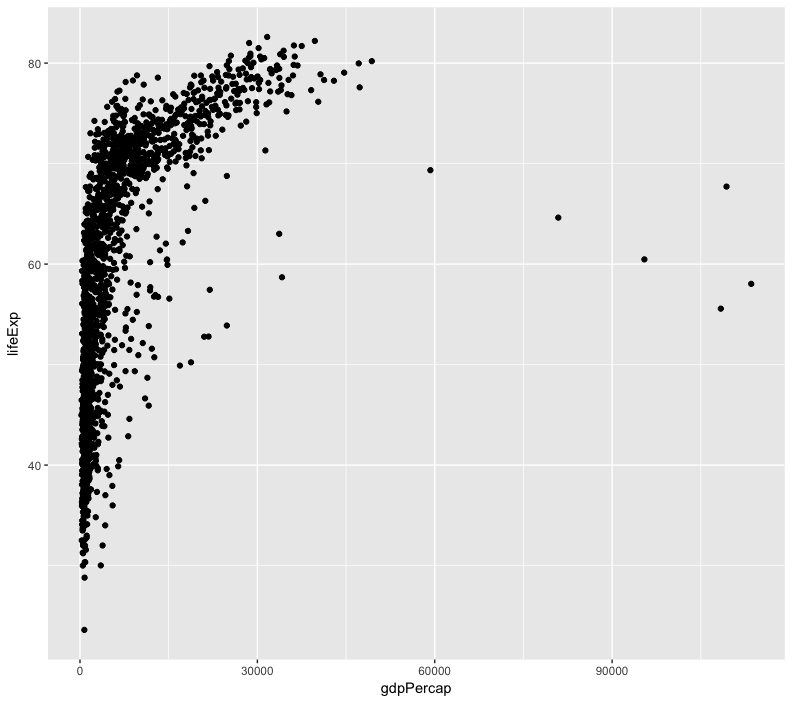
it’s hard to see the relationship between the points due outliers
change the scale of units on the x axis using the scale functions.
can also modify the transparency of the points, using the alpha function - helpful when large amount of data which is very clustered.
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point(alpha = 0.5) + scale_x_log10()
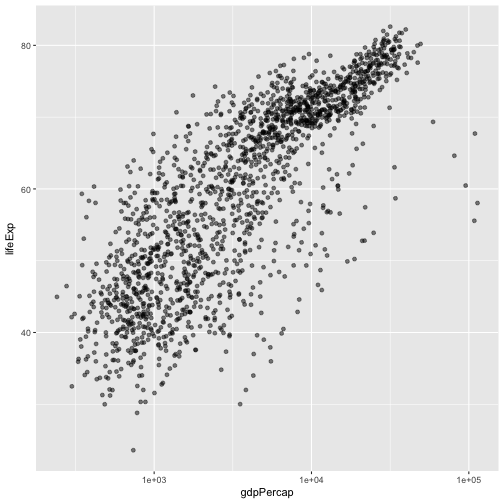
The log10 function applied a transformation to the values of the gdpPercap
column before rendering them on the plot, so that each multiple of 10 now only
corresponds to an increase in 1 on the transformed scale, e.g. a GDP per capita
of 1,000 is now 3 on the y axis, a value of 10,000 corresponds to 4 on the y
axis and so on. This makes it easier to visualize the spread of data on the
x-axis.
point out that the alpha aesthetic is applied only to the point geom. Can also make transparency based on variables such as:
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point(aes(alpha = continent)) + scale_x_log10()
We can fit a simple relationship to the data by adding another layer,
geom_smooth:
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point() + scale_x_log10() + geom_smooth(method="lm")
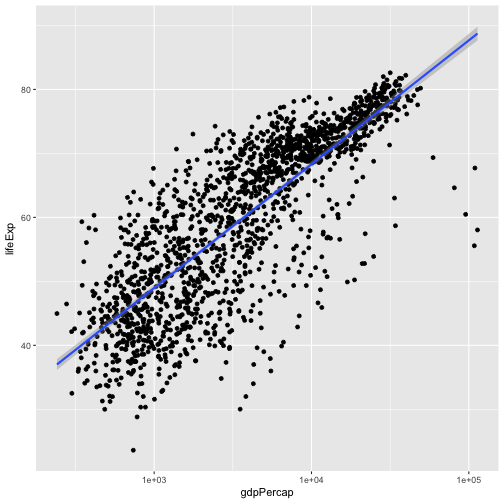
We can make the line thicker by setting the size aesthetic in the
geom_smooth layer:
ggplot(data = gapminder, aes(x = gdpPercap, y = lifeExp)) +
geom_point() + scale_x_log10() + geom_smooth(method="lm", size=1.5)
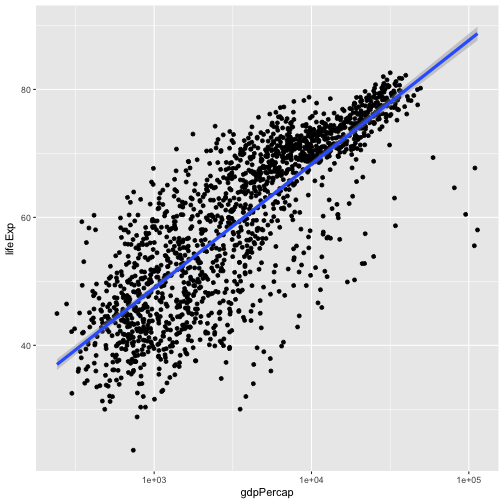
There are two ways an aesthetic can be specified. Here we set the size
aesthetic by passing it as an argument to geom_smooth. Previously in the
lesson we’ve used the aes function to define a mapping between data
variables and their visual representation.
CHALLENGES - 10 min
Multi-panel figures
we can split this out over multiple panels by adding a layer of facet panels. Focusing only on those countries with names that start with the letter “A” or “Z”.
start by subsetting the data
use the substr function to
pull out a part of a character string
the %in% operator allows us to make multiple comparisons rather
than write out long subsetting conditions
starts.with <- substr(gapminder$country, start = 1, stop = 1)
az.countries <- gapminder[starts.with %in% c("A", "Z"), ]
ggplot(data = az.countries, aes(x = year, y = lifeExp, color=continent)) +
geom_line() + facet_wrap( ~ country)
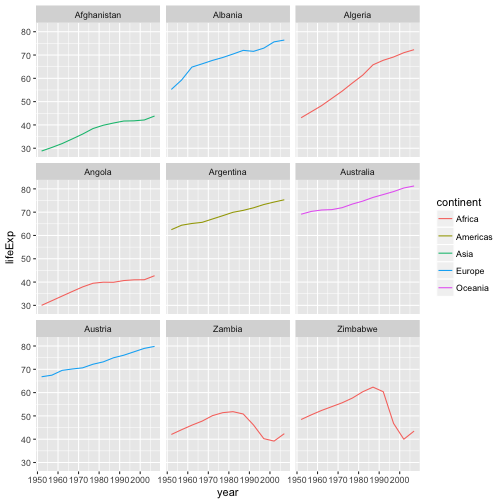
The facet_wrap layer takes a formula as its argument, denoted by the tilde
(~). This tells R to draw a panel for each unique value in the country column
of the gapminder dataset.
Modifying text
change some of the text elements.
rename our x and y axes using the xlab() and ylab() functions:
ggplot(data = az.countries, aes(x = year, y = lifeExp, color=continent)) +
geom_line() + facet_wrap( ~ country) +
xlab("Year") + ylab("Life Expectancy")
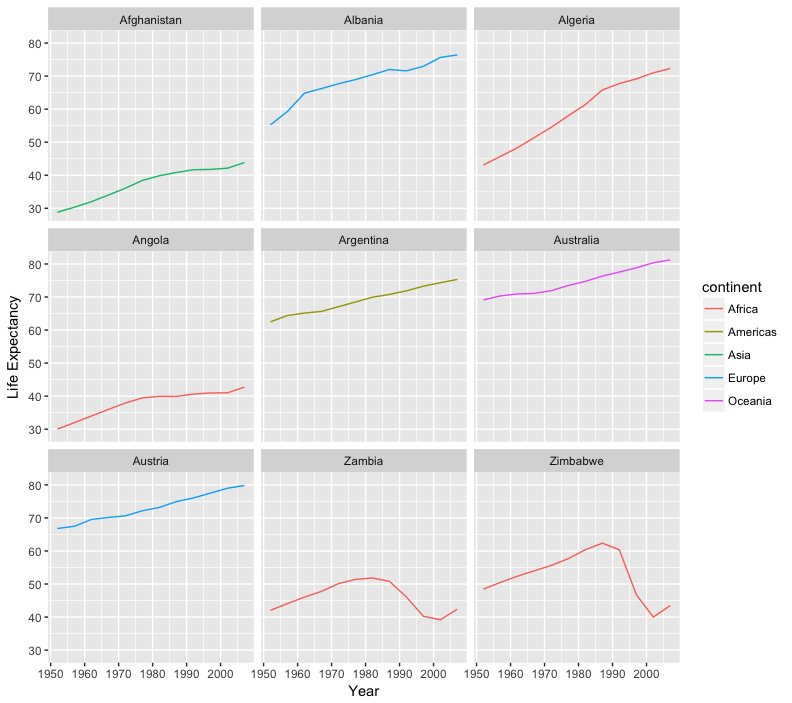
give our figure a title with the ggtitle() function. And capitalize the label of our
legend. This can be done using the scales layer.
ggplot(data = az.countries, aes(x = year, y = lifeExp, color=continent)) +
geom_line() + facet_wrap( ~ country) +
xlab("Year") + ylab("Life Expectancy") +
ggtitle("Figure 1") + scale_colour_discrete(name="Continent")
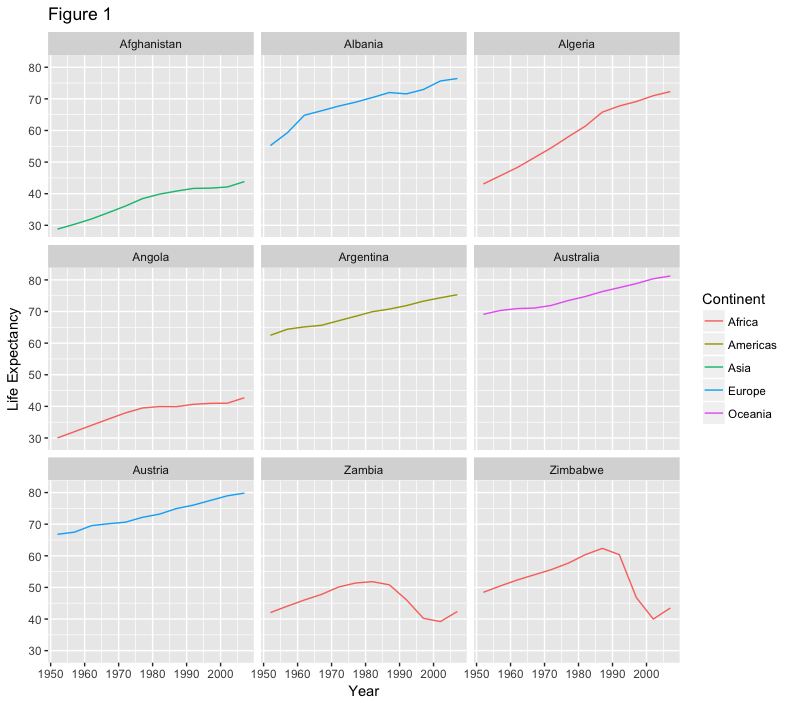
let’s remove the x-axis labels so the plot is less cluttered. To do this, we use the theme layer which controls the axis text and overall text size.
ggplot(data = az.countries, aes(x = year, y = lifeExp, color=continent)) +
geom_line() + facet_wrap( ~ country) +
xlab("Year") + ylab("Life Expectancy") +
ggtitle("Figure 1") + scale_colour_discrete(name="Continent") +
theme(axis.text.x=element_blank(), axis.ticks.x=element_blank())
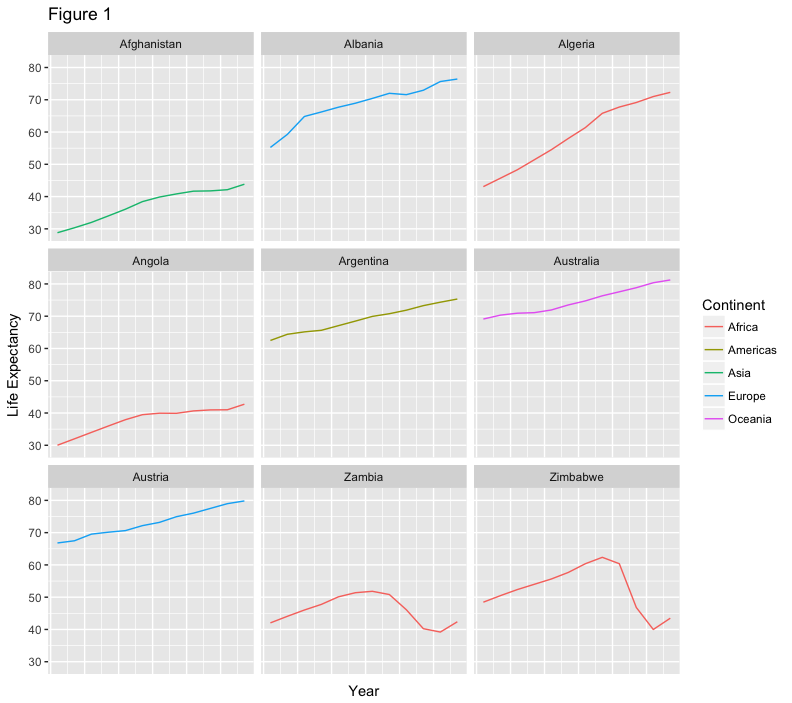
CHALLENGES - allot 5 min
Writing Data - complete by 4:20 pm
Saving plots
So making publication quality plots is great but does us little good if we cannot get them out of R and into our documents.
You can save a plot from within RStudio using the ‘Export’ button in the ‘Plot’ window
if you are working in a command line environment, or want to create multiple plots without user interaction?
you can use the ggsave function to save your plots quickly. This function can be used to save the last displayed plot in a format specified by the file name.
You can save as several different formats, such as a PDF:
ggsave("My_most_recent_plot.pdf")
Or as a JPG:
ggsave("My_most_recent_plot.jpg")
ggsave also allows you to specify size and quality of the image. You can check out all of the
options using the ?ggsave command to view the help file.
if you will want to save plots without creating them in the ‘Plot’ window first. Perhaps you want to make a pdf document with multiple pages: each one a different plot, for example. Or perhaps you’re looping through multiple subsets of a file, plotting data from each subset, and you want to save each plot, but obviously can’t stop the loop to click ‘Export’ for each one.
The function
pdf creates a new pdf device. You can control the size and resolution
using the arguments to this function.
pdf("Life_Exp_vs_time.pdf", width=12, height=4)
ggplot(data=gapminder, aes(x=year, y=lifeExp, color=continent)) +
geom_point()
dev.off()
The pdf command opens the pdf file, and any output between this command and the dev.off() command
will be added to that file.
dont forget the dev.off command
Open up this document and have a look.
CHALLENGES - 5 min
The commands jpeg, png etc. are used similarly to produce
documents in different formats.
Writing data
At some point, you’ll also want to write out data from R.
We can use the write.table function for this, which is
very similar to the read.table function that we mentioned previously in Lesson 2.
Let’s create a data-cleaning script, for this analysis, we only want to focus on the gapminder data for Australia:
make a script file
aust_subset <- gapminder[gapminder$country == "Australia",]
write.table(aust_subset,
file="gapminder-aus.csv",
sep=","
)
in line comments and linebreaks within function calls
aust_subset <- gapminder[gapminder$country == "Australia",]
write.table(aust_subset, ## Gapminder data for countries located in Australia
file="gapminder-aus.csv", ## Name of the output file
sep="," ## Comma separated
)
open a shell window and navigate to the location of the file using your cd command.
to find out where R is saving
your files, you can check the file pane or use the getwd() command.
head gapminder-aus.csv
you can view
the file in R by clicking on the filename in the file pane and selecting View File.
Where did all these quotation marks come from? Also the row numbers are meaningless. let’s look at the help file
?write.table
By default R will wrap character vectors with quotation marks when writing out to file. It will also write out the row and column names.
write.table(aust_subset, ## Gapminder data for countries located in Australia
file="gapminder-aus.csv", ## Name of the output file
sep=",", ## Comma separated
quote=FALSE, ## Turn off quotation marks
row.names=FALSE ## No row names
)
look at the data again using our shell skills:
head gapminder-aus.csv
That looks better!
CHALLENGES - 5 min
Wrap Up - 5 min
Help Files in R
Don’t forget your R helpfiles and package vignettes which can be accessed
by using the ? and vignette commands.
Supplemental Lessons
Additional R topics that we could not cover today.
R Club at UNL
The R Club meets on East Campus twice a month. It is headed by Leonardo Bastos, a PhD student in Agronomy and Horticulture. You can email Leonardo for more information, or check out the club’s GitHub page which contains previous meeting topics.
RStudio cheat sheets
R quick reference guides including today’s handouts and more!
R for Data Science
Hadley Wickham is RStudio’s Chief Data Scientist and developer of the dplyr and ggplot2 packages.
R for Data Science is his newest book, and is available here for free.
One R Tip a Day on Twitter
Following One R Tip a Day is a great way to learn new tips and tricks in R.
Twotorials
Twotorials is a compilation of 2 minute youtube videos which highlight a specific topic in R.
Quick R Website
Cookbook for R
Advanced R
For more advanced topics, check out Hadley Wickham’s website based on his book “Advanced R”.
Remind them to fill out post-it survey